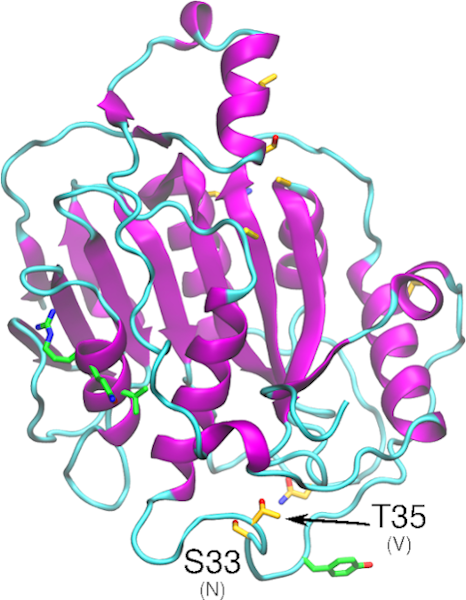
Download The nonstructural protein 13 (nsp13) is involved in a number of functions, such as NTPase, dNTPase, RTpase, RNA helicase, and DNA helicase activity. A single mutation in SARS-CoV-1 nsp13 stalk domain, resulted in a dramatic decrease in viral infectivity in vitro. The essential nature of nsp13 in the viral replication cycle, and its multifunctional […]
Category: COVID-19 Models
Nonstructural protein 15 – endoRNAse
Download Nsp15 is a Nidoviral RNA uridylate-specific endoribonuclease and its C-terminal is a catalytic domain belonging to the EndoU family of enzymes. Within SARS-CoV-1 and -CoV-2, nsp15 is very conserved (89 % identity). Narrative Nsp15 is a Nidoviral RNA uridylate-specific endoribonuclease (NendoU) and its C-terminal is a catalytic domain belonging to the EndoU family of […]
Nonstructural protein 16 – 2’-O-ribose methyltransferase
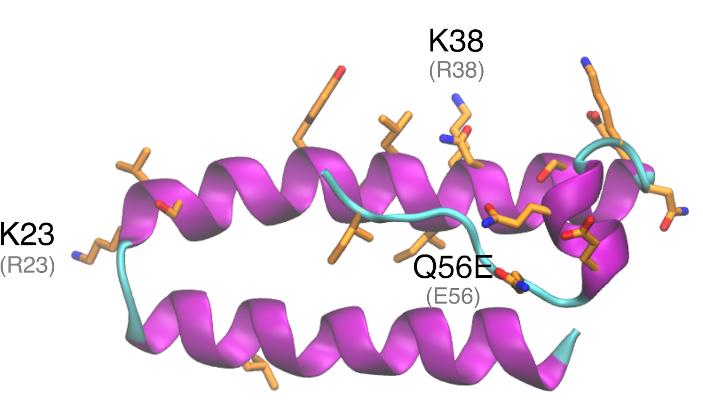
Download The nonstructural protein 16 (nsp16) is involved in capping of viral mRNA to protect it from host degradation, and it has been demonstrated that it has to be associated with nsp10 to be active. Mutation that increases hydrophobic interaction between nsp10 and the hydrophobic pocket of nsp16, increases methyltransferase activity of nsp16. Conversely, […]
Spike glycoprotein
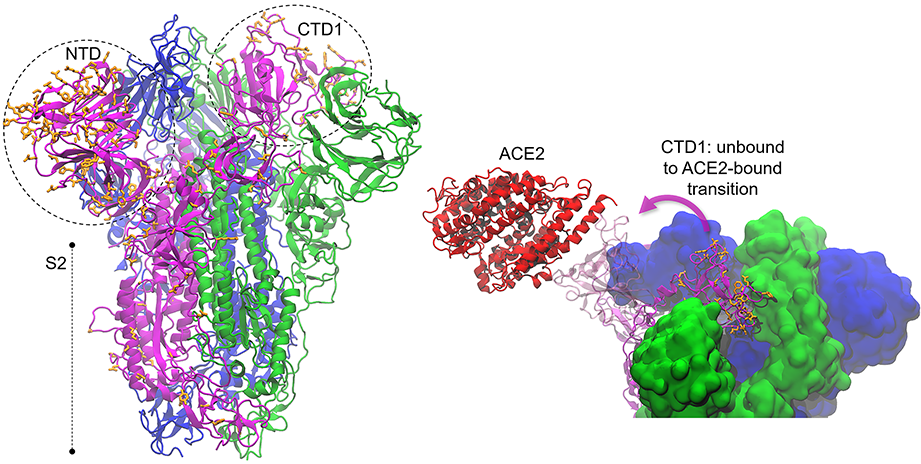
Sequence Analysis Download Simulations ACE2_RBD2_A ACE2_RBD2_B ACE2_RBD2_C ACE2_RBD2_D ACE2_RBD2_E ACE_RBD2_A ACE_RBD2_B ACE_RBD2_C ACE2_RBD1_A ACE2_RBD1_B ACE2_RBD1_C ACE2_RBD1_D ACE2_RBD1_E ACE_RBD1_A ACE_RBD1_B ACE_RBD1_C The spike glycoprotein (S) is the main promoter of virus entry into host cells. These highly glycosylated proteins protrude from the viral surface to interact with the host cell receptor(s). The receptor binding domain of […]
ORF3a
Download ORF3a is an integral transmembrane protein, localized mostly in the Golgi complex, cytoplasm and cell surface. ORF3a is able to form homotetramers with ion channel properties. Regarding pathogenic effects, ORF3a is linked to inflammatory responses, weak INF responses, innate immunity responses, trigger apoptosis and modulate cell cycle. Non-conservative mutations with a potential impact in […]
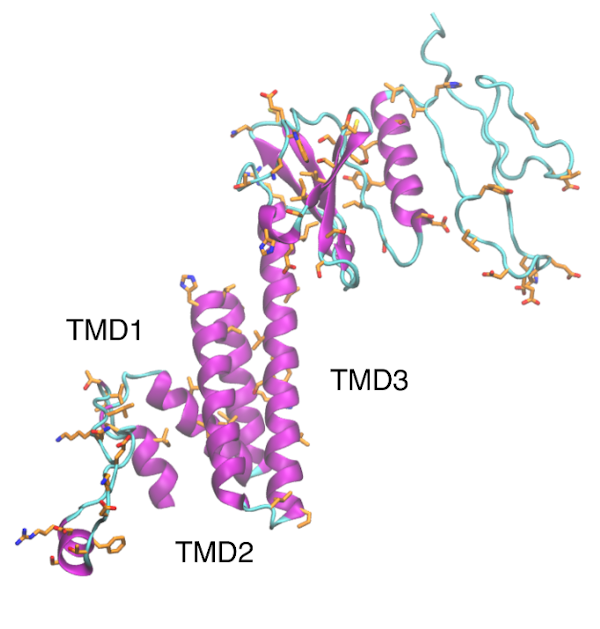
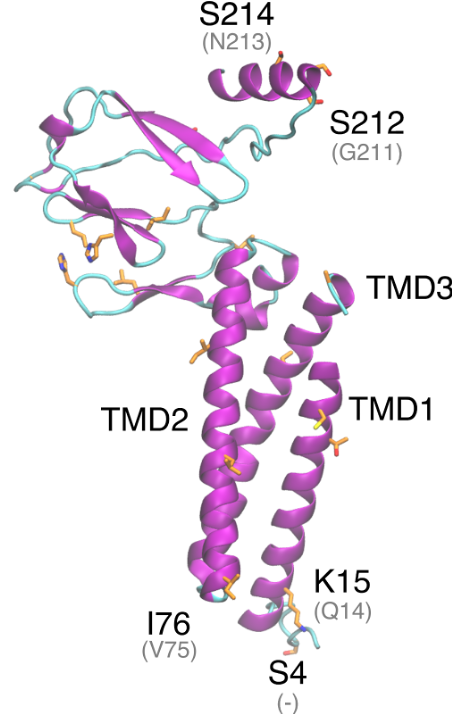
Envelope protein
Download E protein is a type 1 transmembrane protein able to form pentamers creating pores with ion transport activity. E protein localizes mainly in the ER and Golgi apparatus, where it participates in assembly, budding and intracellular trafficking of newly formed virions. E protein is involved in tight junctions disruption in the lungs to reach […]
Membrane protein
Download Membrane (M) protein is the most abundant of all coronavirus structural proteins. M protein main functions are membrane curvature initiation, RNA packing and viral particle budding. Besides, M protein has been linked to apoptosis, suppression of inflammatory responses and IFN signaling. Most of the variation between SARS-CoV and SARS-CoV-2 M occurs in the cytoplasmic […]
ORF6
Download ORF6 localizes in the perinuclear and endoplasmic reticulum. ORF6 is known to interact with the viral proteins ORF9b and nsp8, suggesting ORF6 may play a role in virus replication. Besides, ORF6 has been associated with impairing the activation of IFN signaling and membrane rearrangements. SARS-CoV-1 and SARS-CoV-2 ORF6 proteins are 68.85% identical, with most […]
ORF7a
Download ORF7a is a type I transmembrane protein, localized mainly in ER, Golgi apparatus and in the cell surface. ORF7a is able to prevent virus tethering at the plasma membrane by BTS-2. Further in vitro experiments showed that ORF7a is able to induce apoptosis in a caspase dependent manner. ORF7a protein is very conserved between […]
ORF7b
Download This accessory protein is an integral transmembrane protein and it localizes in the cis- and trans- Golgi. MSARS-CoV-2 ORF7b is 81 % identical to SARS-CoV-1 ORF7b. All substitutions are found in the terminals, being the transmembrane helix fully conserved. Narrative ORF7b is a small accessory protein expressed in SARS-CoV-1 and SARS-CoV-2 infected cells with […]