ORF6
ORF6 localizes in the perinuclear and endoplasmic reticulum. ORF6 is known to interact with the viral proteins ORF9b and nsp8, suggesting ORF6 may play a role in virus replication. Besides, ORF6 has been associated with impairing the activation of IFN signaling and membrane rearrangements. SARS-CoV-1 and SARS-CoV-2 ORF6 proteins are 68.85% identical, with most of the substitutions located in the C-terminus helix. Among them, the introduction of new putative ubiquitination sites and mutations in a critical area for ORF6 function were verified.
Narrative
ORF6 is an auxiliary protein in SARS coronaviruses that is not required for virus replication (Yount et al. 2005; C. Huang, Peters, and Makino 2007). However, it can increase virus replication when expressed in a heterologous system or at low multiplicity of infection (Zhao et al. 2009). ORF6 localizes in the perinuclear and ER zones, associated with membranes and colocalized with M , S, and N structural proteins (Geng et al. 2005; Pewe et al. 2005). ORF6 is known to interact with the viral proteins ORF9b and nsp8 (Kumar et al. 2007; Calvo et al. 2012). Furthermore, ORF6 and nsp8 colocalize in cell culture, suggesting ORF6 may play a role in virus replication (Kumar et al. 2007).
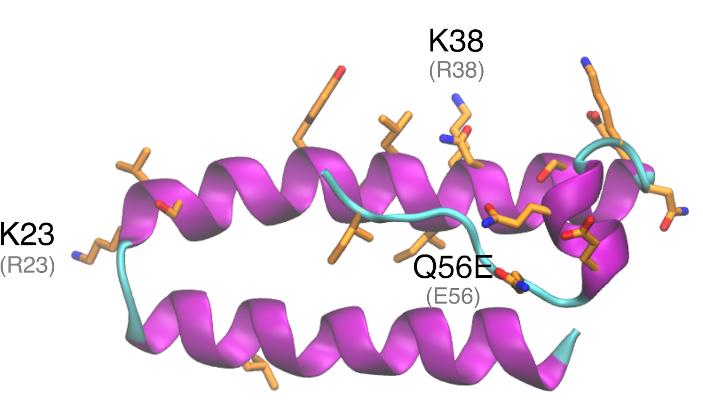
Structural analysis and comparison with SARS-CoV-1 ORF6 – SARS-CoV-1 ORF6 is a small protein of 63 amino acids and 7·53 kDa. Several important residues have been related to SARS-CoV-1 ORF6 structure, function, and location. For instance, ORF6 sequence suggests a membrane association in residues 7 – 37 (Pewe et al. 2005). Even though some residues in this region are charged (Glu13, Arg20 and Lys23, in SARS-CoV-2 ORF6), this was enough for membrane association (Pewe et al. 2005).
ORF6 prevents import of the host signal transducer and activator of transcription 1-alpha/beta (STAT1) to the nucleus by tethering karyopherin subunit alpha-2 (KPNA2) and, therefore, KPNA1 to the ER and golgi apparatus membrane, thereby impairing the activation of INF-induced genes (Frieman et al. 2007). Tests with specific mutations in the C-terminal region of ORF6 revealed specific residues that affect disruption of KPNA2 ER and Golgi arrest in ORF6 transfected cells (Frieman et al. 2007). Further, the ORF6 C-terminal was shown to interact with Nmi protein, mediating Nmi ubiquitination and proteasome degradation, thus suppressing INF signaling (Cheng et al. 2015). ORF6 N-terminal, in turn, was shown to induce membrane rearrangements typically observed in virus-infected cells and to have critical importance to prevent STAT1 translocation to the nucleus (Zhou et al. 2010).
SARS-CoV-1 and SARS-CoV-2 ORF6 proteins are 68·85% identical. Most of the substitutions are located in the C-terminal helix. Lysine substitutions Arg23Lys, Arg38Lys and Asn38Lys in SARS-CoV-2 ORF6 suggest the introduction of new putative ubiquitination sites. ORF6 has been shown to be involved in proteasomal degradation of Nmi, but if ORF6 itself is regulated by the proteasome system is unknown.
Two studies pointed to residues 53 – 63 to be important for SARS-CoV-1 ORF6 function (Zhou et al. 2010; Frieman et al. 2007). In this region, one non-conservative substitution (Glu56Gln) and two deletions (Tyr62 and Pro63) are verified.
