Download In vitro analyses have found that SARS-COV-1 nsp1 may disrupt the host interferon defense response, by potentially affecting the downstream defense signaling. The nsp1 protein has been shown to bind the 40S Ribosomal subunit, which has been associated with degradation of host mRNA, and suppression of host mRNA translation, leaving the viral RNA unaffected. […]
Nonstructural Protein 2
Download The specific role of nsp2 has not been established; it may be that nsp2 assists other viral proteins in performing their function, such as regulating the autophagy defense response or promoting mitochondrial dysfunction, thereby helping viral replication or effecting disease severity. Some evidence links nsp2 to mitochondrial dysfunction and autophagy via prohibitin 1 (PHB1), […]
Nonstructural Protein 3 – Papain-like proteinase
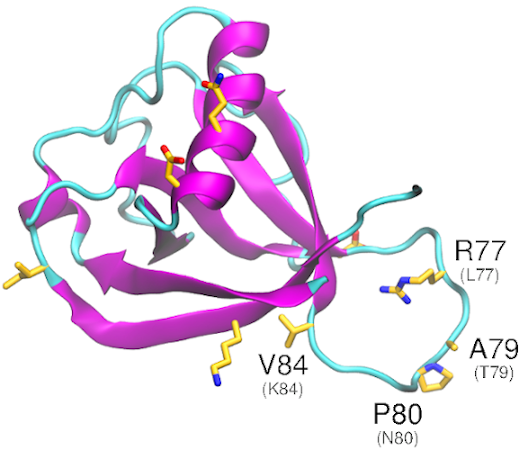
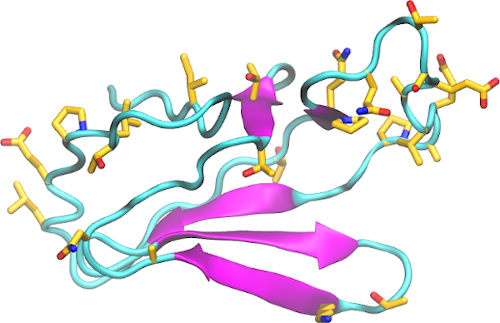
Download The nonstructural protein 3 (nsp3) acts as a phosphatase and its catalytic domain is conserved, sharing homology with yeast, archaea, and E. coli. The papain-like protease domain of nsp3 is implicated in inhibiting components of NF-κB, interferon-beta, and p53, thereby disrupting the host immune response. A substitution in SARS-CoV-2 likely intensifies the interaction with […]
Nonstructural protein 4
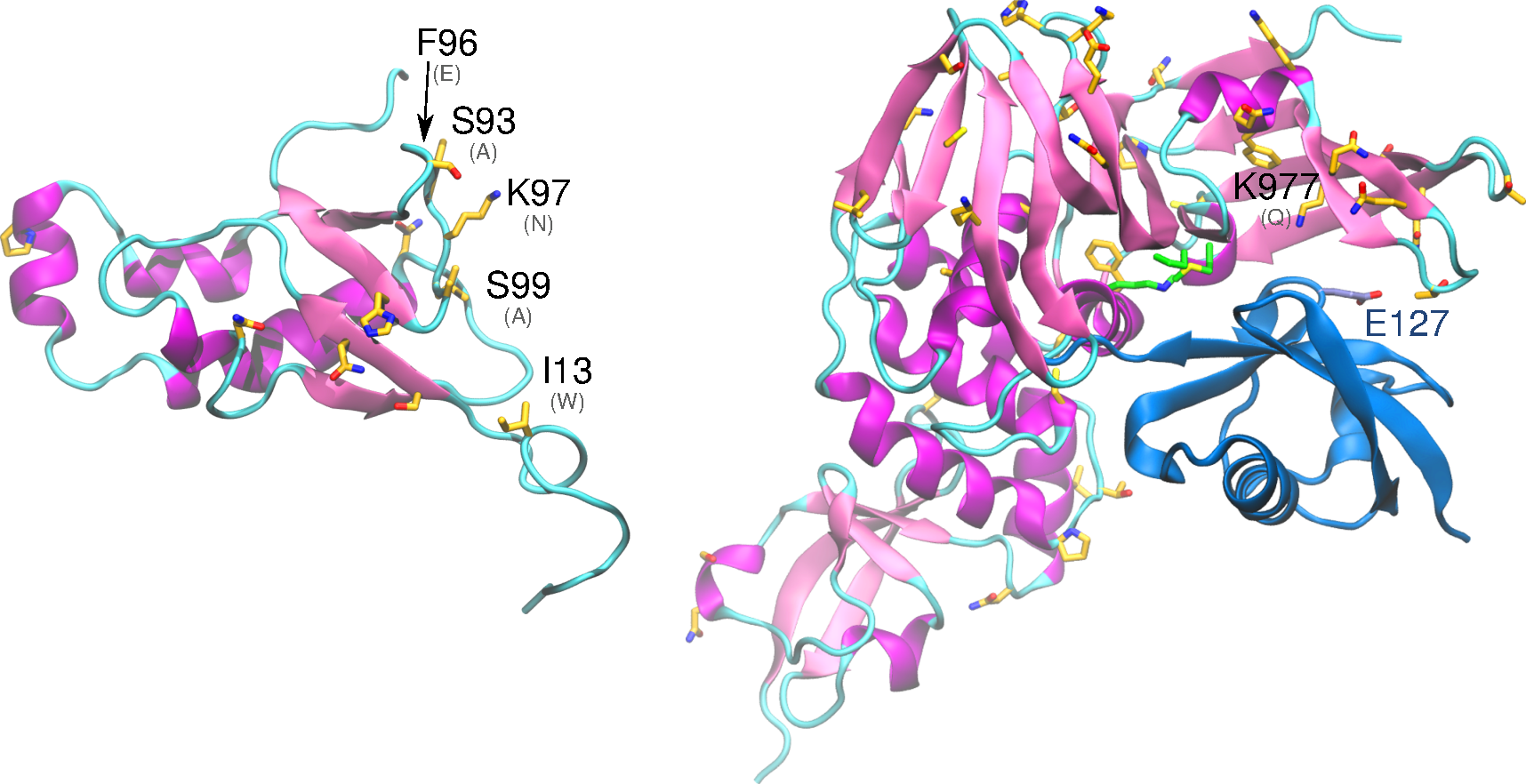
Download The nonstructural protein 4 (nsp4) is essential for membrane rearrangements during viral replication in a mechanism involving nsp3. Nsp4 is thought to be a tetra spanning transmembrane protein and disruption in glycosylation sites within the luminal loop give rise to aberrant double membrane vesicles. Structural information about nsp4 is scarce, thus a low-resolution model […]
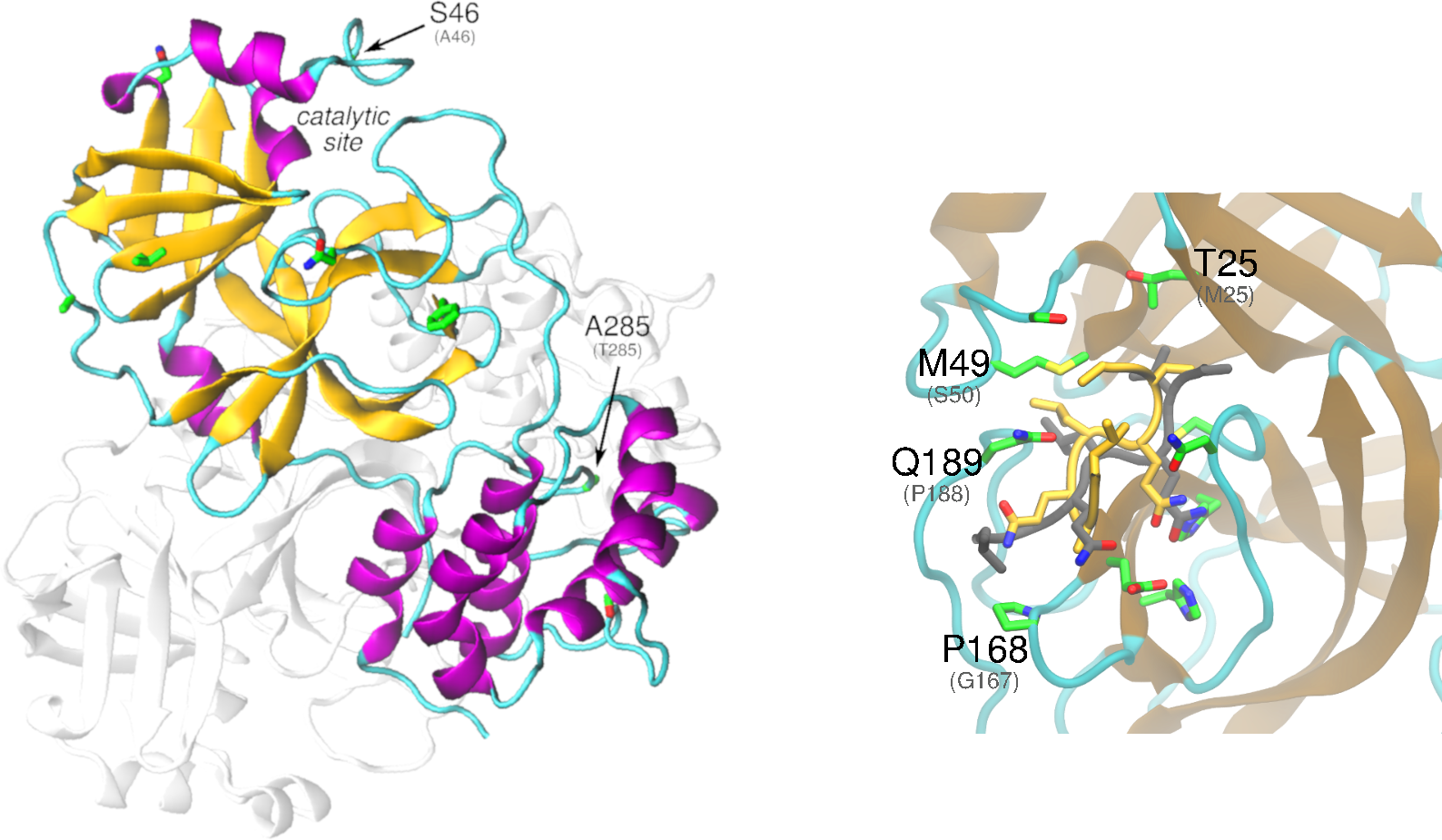
Nonstructural protein 5 – 3C-like proteinase
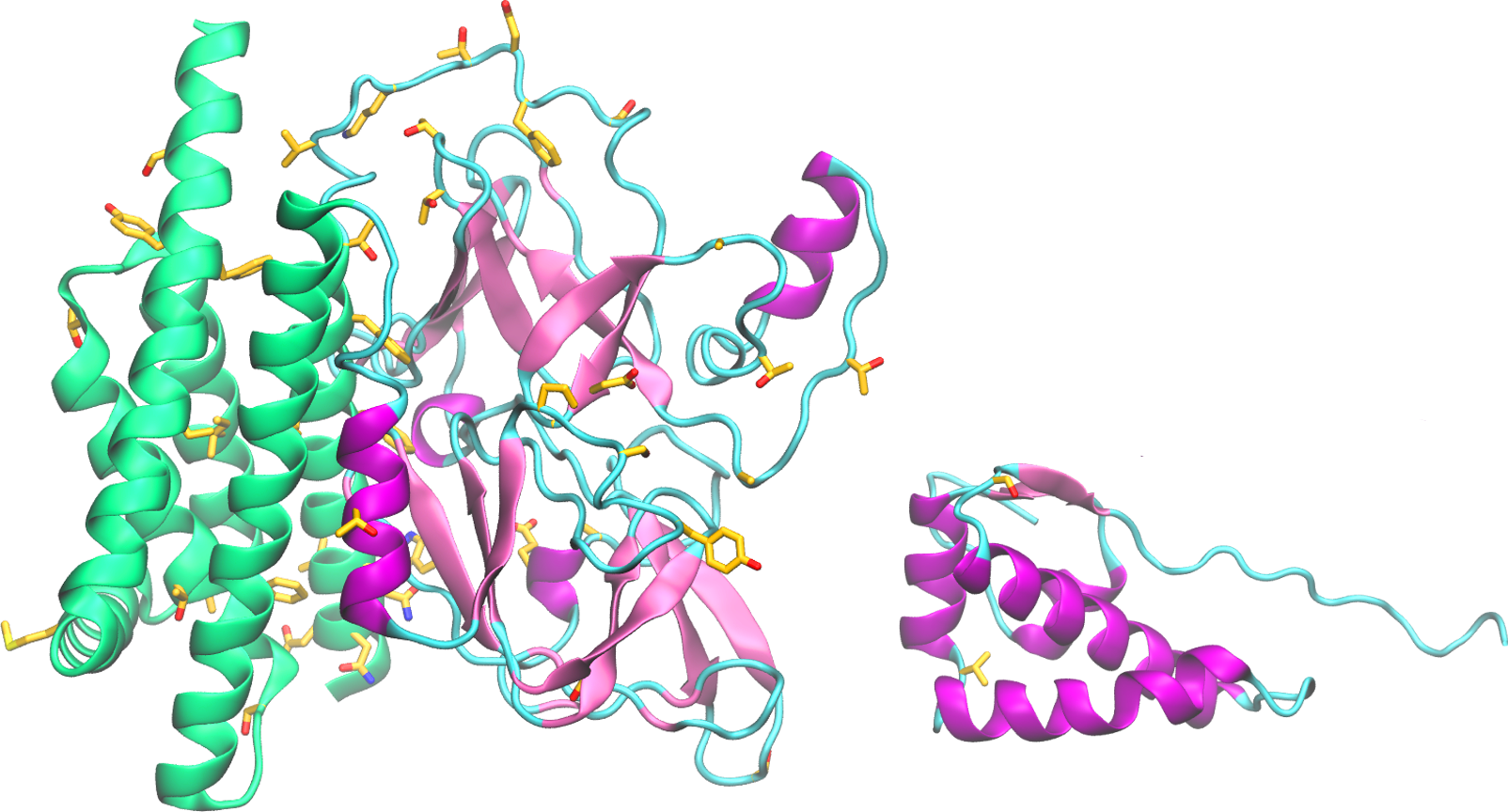
Download The nonstructural protein 5 (nsp5, also known as 3CLpro), is the main protease of the coronavirus genome, exhibiting as main known role the cleavage of the polyproteins translated from the viral RNA viral. Dimerization of SARS-CoV-1 3CLpro is essential to stabilize the catalytic site. The dimer interface is highly conserved within SARS-CoV-1 and -CoV-2, […]