Nonstructural protein 5 – 3C-like proteinase
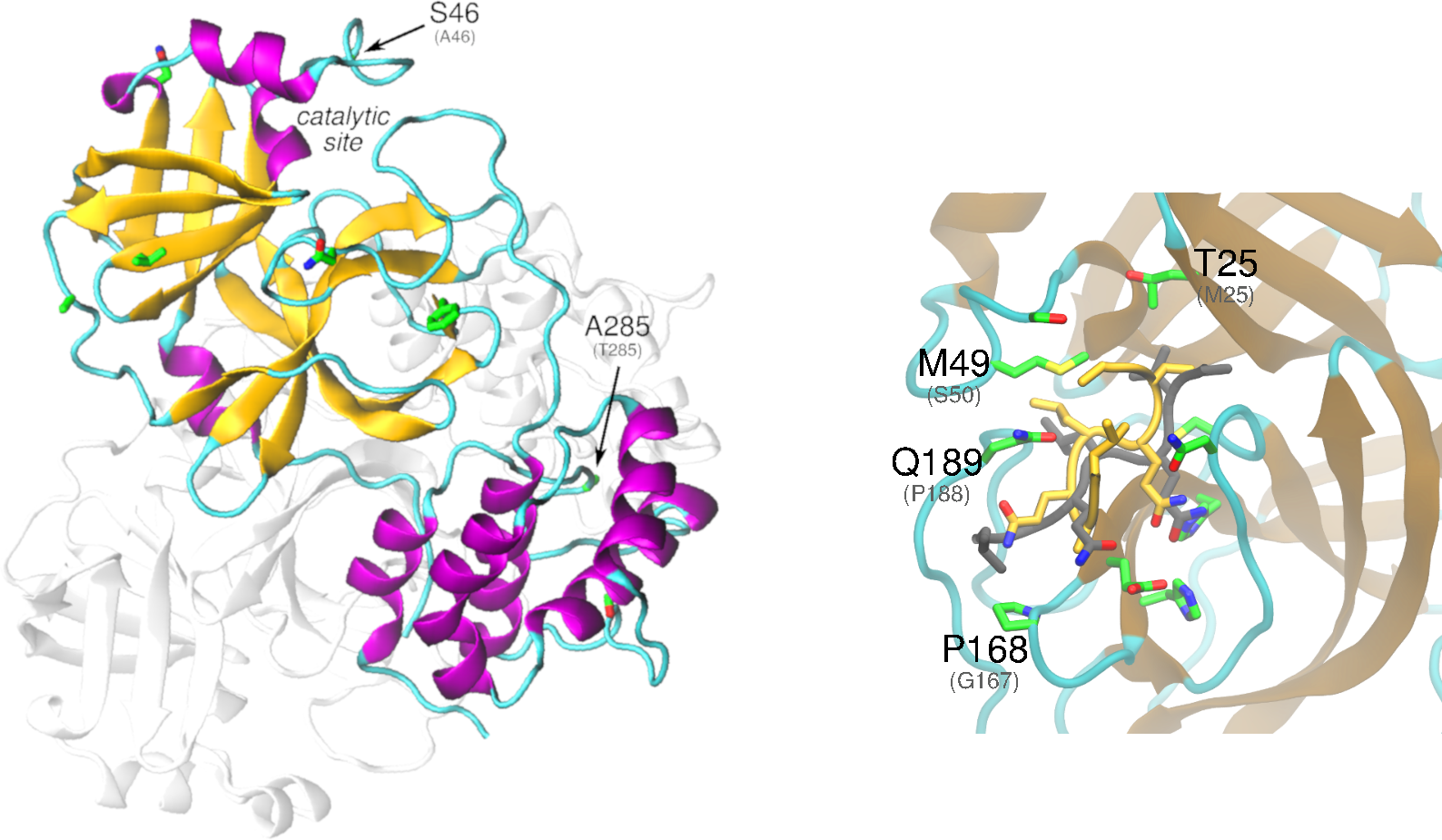
The nonstructural protein 5 (nsp5, also known as 3CLpro), is the main protease of the coronavirus genome, exhibiting as main known role the cleavage of the polyproteins translated from the viral RNA viral. Dimerization of SARS-CoV-1 3CLpro is essential to stabilize the catalytic site. The dimer interface is highly conserved within SARS-CoV-1 and -CoV-2, however, substitutions may affect dimer interaction and phosphorylation pattern of 3CLpro in SARS-CoV-2. Given that the substrate binding site of SARS-CoV-2 3CLpro is very similar to PEDV 3CLpro, it is possible that SARS-CoV-2 3CLpro is also active towards NF-kB essential modulator, thus suppressing host immune response.
Narrative
The nonstructural protein 5 (nsp5, also known as 3CLpro), is the main protease of the coronavirus genome, exhibiting a main role in the cleavage of the polyproteins translated from the viral RNA (Perlman and Netland 2009; Ziebuhr, Snijder, and Gorbalenya 2000; Anand et al. 2003). This protein is highly conserved relative to SARS-CoV-1 (96% identity) and among RNA+ viruses (Nidovirales) in general, making it an attractive target for pan-antiviral drugs (Zhang, Lin, Kusov, et al. 2020; Nukoolkarn et al. 2008; Dayer, Taleb-Gassabi, and Dayer 2017). In addition, it has been shown that loss of nsp3 and nsp10 substantially reduces 3CLpro activity (Donaldson et al. 2007, Stokes et al. 2010) and therefore therapeutics that target these proteins could indirectly inhibit 3CLpro.
Structural analysis and comparison with SARS-CoV-1 nsp5 – Studies with SARS-CoV-1 show that dimerization is essential to stabilize the productive conformation of 3CLpro catalytic site. The recently solved structure of 3CLpro of SARS-CoV-2 (PDB id: 6y2e) confirms the dimer as its biological state. The dimer interface is highly conserved within SARS-CoV-1 and -CoV-2, as well as other residues that indirectly affect dimerization, such as Ser144, Ser 147 and Asn28 (Barrila, Bacha, and Freire 2006). However, a relevant substitution Thr285Ala is found on the interface. Based on previous studies with SARS-CoV-1 3CLpro, this replacement is thought to favor the hydrophobic pack within monomers, and it was recently associated with a slightly higher catalytic efficiency of SARS-CoV-2 compared to SARS-CoV-2 (Zhang, Lin, Sun, et al. 2020). The analysis of the evolutionary tree from the aligned sequences of coronavirus from all available species reveals that alanine at site 285 defines the SARS-CoV-2 clade and three bat coronaviruses from mainland China (manuscript in review). In contrast, the many of the beta coronaviruses that infect mammals have a cysteine at this location. Given the proximity with the cysteine in the opposite monomer, it is possible that a disulfide bridge is formed in these proteases, which may result in a more tightly bound dimer and increased catalytic efficiency. Further exploration of this site is warranted.
There are 7 non-conservative amino acid substitutions between SARS-CoV-1 and SARS-CoV-2, besides Thr285Ala. Most of those are located in regions that are not clearly associated with protease function, except by Ala46Ser, positioned at the catalytic cleft entrance, which may affect substrate affinity/selectivity. Moreover, the phosphorylation of serines in 3CLpro of rotaviruses was shown to be essential for protease activity. The substitutions Ala46Ser, Ser65Asn, Ser94Ala, Ala267Ser and Thr285Ala may also affect the phosphorylation pattern of 3CLpro in SARS-CoV-2.
