Nonstructural Protein 2
The specific role of nsp2 has not been established; it may be that nsp2 assists other viral proteins in performing their function, such as regulating the autophagy defense response or promoting mitochondrial dysfunction, thereby helping viral replication or effecting disease severity. Some evidence links nsp2 to mitochondrial dysfunction and autophagy via prohibitin 1 (PHB1), PHB2 and LC3.
Narrative
Positive RNA viruses create viroplasms, subcellular compartments that are thought to protect the viral machinery against host-defense strategies, and even facilitate replication (Netherton and Wileman 2011). They often contain double stranded RNA, as well as the replication complex of the virus. For SARS-CoV-1, the replicase proteins assemble at the cytosol side of the ER, after which invagination occurs. There is no evidence of a pore maintaining the connection of the invaginated space to the cytosol, thus no maintained sphererule has been observed, though vesicles do form. A network of double-membrane-vesicles (DMVs) is eventually formed, anchored to the ER by a convoluted-membrane (CM) compartment. Replication appears to happen in the CM, while DMVs directly connected to the CM contain replicase proteins and viral proteins. Those DMVs not directly connected to CM form vesicle packets containing viral genomes (Netherton and Wileman 2011; Knoops et al. 2008).
In Hagemeijer et al. (2010), it was shown that the nonstructural protein 2 (nsp2) localized to the membranes of CM and DMV, and it is exposed to the cytoplasm. It appears that during formation of the replication complex, nsp2 is recruited, but once formed no further exchange of nsp2 occurs. Mutants of SARS-CoV-1 with the deleted nsp2 coding sequence are still capable of viral replication, although with decreased growth (Graham et al. 2005). The specific role of nsp2 has not been established; it may be that nsp2 assists other viral proteins in performing their function, such as regulating the autophagy defense response or promoting mitochondrial dysfunction, thereby helping viral replication or effecting disease severity. It has been shown that nsp2 interacts directly with prohibitin 1 (PHB1) and PHB2 (Cornillez-Ty et al. 2009) host proteins with a wide variety of functions. Kathiria et al. (2012) have shown that PHB knockdown resulted in increased ROS, mitochondrial depolarization, and induced autophagy. In (Hernando-Rodríguez and Artal-Sanz 2018) several phenotypes of PHB are reviewed, including the role of PHB in mitochondrial stability. PHB has also been implicated in inflammatory response in both the lung and gut (Hernando-Rodríguez and Artal-Sanz 2018; Agrawal, Gupta, and Agrawal 2012). In von Brunn et al. (2007), it was shown that nsp2 displayed co-immunoprecipitation (CoIP) interaction with N-terminal side of nsp3, nsp6, nsp8, nsp11, nsp16 and ORF3a, and co-localization with nsp8 and nsp3, where nsp8 almost always co-localized with the microtubule protein, LC3, an autophagy marker protein (Prentice et al. 2004). These results suggest that nsp2 may be involved in mitochondrial dysfunction through PHB1 and 2, and association with LC3 may be worth investigating. Another potential for nsp2’s association with mitochondrial dysfunction may lie with its CoIP interaction with ORF3a, as the SARS protein ORF3a is known to disrupt mitochondrial stability (Padhan et al. 2008).
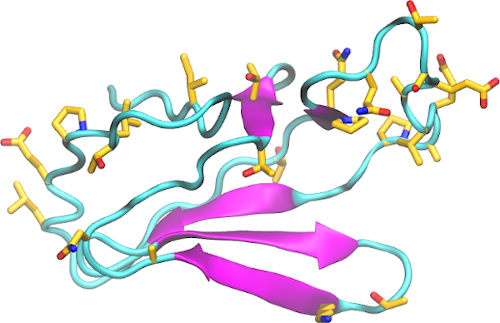
Structural analysis and comparison with SARS-CoV-1 nsp2 – Structure information about nsp2 is scarce. SARS-CoV-2 nsp2 is 68% identical to SARS-CoV-1 nsp2. Among the substitutions, 130 are non-conservative and highly concentrated in the C-terminal end of the protein. An ab initio model was generated for this region (556-633, figure). Fewer mutations appear at the N-terminal side, and a highly conserved region appears in the middle of the protein sequence.
