Nonstructural Protein 3 – Papain-like proteinase
The nonstructural protein 3 (nsp3) acts as a phosphatase and its catalytic domain is conserved, sharing homology with yeast, archaea, and E. coli. The papain-like protease domain of nsp3 is implicated in inhibiting components of NF-κB, interferon-beta, and p53, thereby disrupting the host immune response. A substitution in SARS-CoV-2 likely intensifies the interaction with host interferon-stimulated gene products.
Narrative
The nonstructural protein 3 (nsp3) acts as a phosphatase and its catalytic domain is conserved, sharing homology with the Ymx7 protein in yeast, AF1521 in the archaea (Archeoglobus fulgidus), and Er58 in E. coli (Saikatendu et al. 2005). The protein comprises at least six domains: 1) an N-terminal ubiquitin-like (Ubl1) domain followed by glu-rich acidic domain; 2) an X domain with a predicted Appr-100-p processing activity; 3) a SUD domain (SARS-specific unique domain); 4) a peptidase C-16 domain that contains the papain-like protease (abbreviated PLnc); 5) a transmembrane domain; and 6) the Y domain.
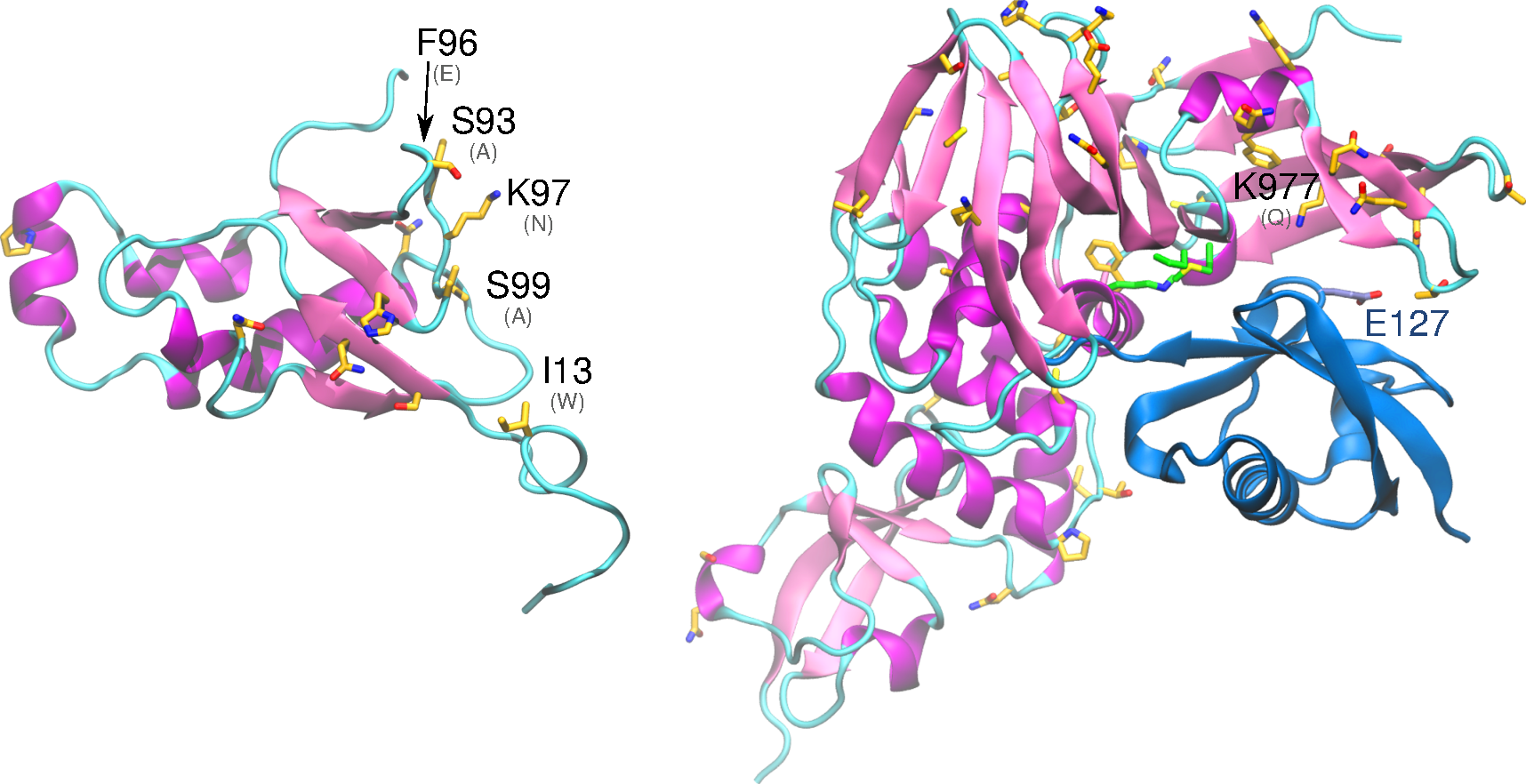
Structural analysis and comparison with SARS-CoV-1 nsp3 – Ub1 binds the nucleocapsid (N) protein and the interaction involves acidic residues of Ubl1 helix α2 and the serine and arginine rich region of the N protein (Lei, Kusov, and Hilgenfeld 2018). Ub1 also binds to the 5’ untranslated region of coronavirus RNA. Ub1 has significant structural homology with Ras effector proteins, and it is thought that Ub1 may interact with and modulate the activity of Ras in the host. This interaction potentially affects growth and cell cycle cascades and could be a potential mechanism for cell mortality (Serrano et al. 2007). Significantly, there are differences in residue identity between SARS-CoV-1 and SARS-CoV-2 in residues 41 to 63, 83 to 87, 88 to 94, 95 to 98 of the Ub1 region (figure, left). These regions correspond to structural homology with Ras effectors and these residue changes may affect interactions with host Ras and contribute to pathogenicity divergence (Serrano et al. 2007).
The glu-rich acidic region is also known as the hypervariable region (HVR). It is intrinsically disordered and its interaction partners are not entirely clear. One study by yeast-2-hybrid demonstrates interaction with nsp6, but a GST pull-down assay also revealed interactions with nsp8, nsp 9, and 3 regions of nsp3 itself (nucleic-acid binding domain, betacoronavirus-specific marker domain, and transmembrane region 1) (Lei, Kusov, and Hilgenfeld 2018). This region is significantly elongated, relative to the SARS-CoV-1 counterpart, with 16 additional amino acids, including several potential sites of glycosylation.
The X domain binds ADP-ribose, the structure of this complex was recently solved for SARS-CoV-2 (PDB id 6w02), and there are several studies and structures available in the Protein Data Bank of homolog complexes (PDB id: 5hol, 5dus, and 2fav). ADP-ribosylation is a type of post-translational modification (PTM), which may be implicated in inhibiting the immune response of the host through PTM of proteins related to expression of interleukin-6 and interferon-beta (Lei, Kusov, and Hilgenfeld 2018). A GST-pulldown assay showed interaction of the X domain of nsp3 with nsp12. Several non-conservative substitutions occur in this domain relative to SARS-CoV-1, but those are located in superficial regions and do not significantly affect protein conformation (RMSD 1.5 Å, relative to 2fav). The papain-like protease domain (papain-like protease 2; PL2pro) (Lei, Kusov, and Hilgenfeld 2018) contains two ubiquitin binding sites. It is implicated in suppression of the host immune response, but its targets and, more generally, which immune-related signal transduction cascades are affected is unclear. It is implicated in inhibiting components of NF-κB, interferon-beta, and p53. In a structural study, PL2pro was found to bind ubiquitin-like interferon-stimulated gene product 15 (ISG15) (Daczkowski et al. 2017), the latter an important post-translational modification of host proteins, including cytokines like interferon. It is believed that cleaving these posttranslational modifications of cytokine proteins by PL2pro disrupts the host immune response (Daczkowski et al. 2017). Importantly, ISG15 has significant interspecies variability, potentially contributing to its very different virulence patterns among species. This region is mostly conserved within SARS-CoV-1 and SARS-CoV-2, including residues that were identified as critical for the interaction with ISG15, namely Arg911, Met953 and Pro992. However, the substitution Gln977Lys likely intensifies the interaction with ISG15 by forming a salt bridge with Glu127, suggesting an important mechanism for variable virulence (figure, right).
