Nucleocapsid protein
Nucleocapsid (N) protein packages the viral RNA into a helical ribonucleocapsid and plays a key role in viral assembly. SARS-CoV-2 N protein is a 419 amino acid long protein and is 89% identical to SARS-CoV-1 nucleocapsid. The RNA binding domain of N is located at the N-terminal and all these residues are fully conserved in SARS-CoV-2 N.
Narrative
Highly abundant in infected cells, SAR-CoV-1 nucleocapsid (N) packages the viral RNA into a helical ribonucleocapsid and plays a key role in viral assembly (Chang et al. 2009). The N protein is assembled and organized in a modular fashion and has been shown to bind to viral RNA at multiple sites in an allosteric manner (Chang et al. 2014). The modularity of the N protein is thought to increase selectivity through coupled allosteric binding of individual nucleocapsid domains, aid in regulation and functional expression, and independently evolve binding sites (Chang et al. 2014).
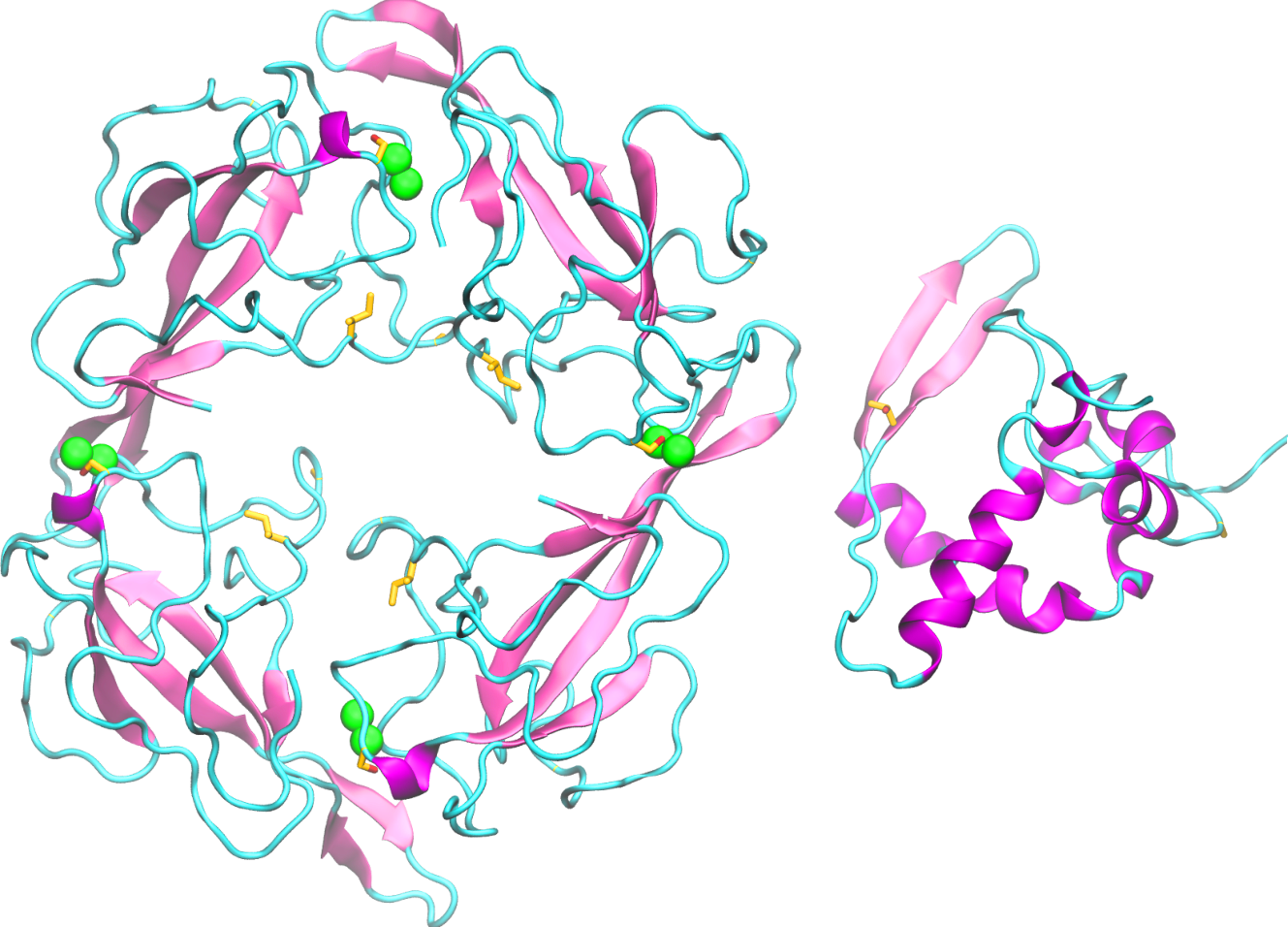
Structural analysis and comparison with SARS-CoV-1 nucleocapsid – SARS-CoV-2 N protein is a 419 a.a. long protein and is 89% identical to SARS-CoV-1 nucleocapsid. In SARS-CoV-1, a Ser-Arg rich motif (Ser-Ser-Arg-Ser-Ser-Ser-Arg-Ser-Arg-Gly-Asn-Ser-Arg) was found to be important for the oligomerization of N proteins (He, Dobie, et al. 2004). This motif is mostly conserved in SARS-CoV-2, except by the substitutions Gly192Asn and Asn193Ser. The RNA binding domain of the N protein is located at the N-terminal, within residues 47 and 180 of SARS-CoV-2. The structure of this domain, recently solved (PDB 6vyo). Residues within the N-terminal of the SARS-CoV-1 nucleocapsid, namely, Tyr87, Tyr110, Tyr112, Y113, Leu122, and Ala13, are hypothesized to play a role in ribonucleocapsid packaging (Chang et al. 2014). All of these residues are fully conserved in SARS-CoV-2 N. A long intrinsically disordered region follows these residues (181-246), and is predicted to fold upon binding (ANCHOR2 prediction (Mészáros, Erdos, and Dosztányi 2018)).
The oligomerization domain of nucleocapsids is situated in the C-terminal of SARS-CoV-1 nucleocapsid. Residues Trp302, Ile305, Pro310, Phe315, and Phe316 are thought to be associated with highly hydrophobic interactions between two helices within the nucleocapsid (Chang et al. 2014), and they are fully conserved in SARS-CoV-2 N.
