Nonstructural protein 9
Nsp9 is able to form dimers that bind either ssDNA and ssRNA and is thought to protect the coronavirus genome from degradation during replication. Deletion of nsp9 in the mouse β-coronavirus (hepatitis virus , MHV), impairs viral RNA synthesis and viral infection. SARS-CoV-1 and SARS-CoV-2 nsp9 sequences are highly conserved, but substitutions with potential impact in post-translational modifications were verified.
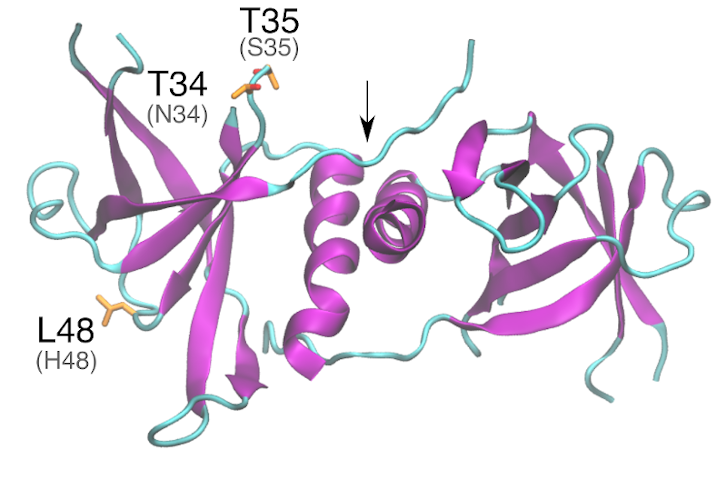
Structural analysis and comparison with SARS-CoV-1 nsp9 – Overall, the structure of nsp9 is very well conserved among β-coronaviruses (Z. Zeng et al. 2018; Sutton et al. 2004). In SARS-CoV-1, nsp9 consists of a small globular protein (113 amino acid residues) with seven antiparallel β-sheets and one α-helix (Sutton et al. 2004). Parallel dimers occur between two nsp9 monomers interacting in their C-terminal α-helixes and their N-fingers (Z. Zeng et al. 2018). In SARS-CoV-1, Gly100 and Gly104 residues are shown to be in the core of the dimer interface (Sutton et al. 2004). Mutation of these conserved glycines in the nsp9 α-helix impairs ssDNA binding, suggesting that dimerization is essential for nucleic acid binding (Z. Zeng et al. 2018). Site-specific mutagenesis studies in the alpha-coronavirus Porcine Epidemic Diarrhea (PEDV) nsp9 identified other key residues for dimerization. The substitutions Lys10Ala, Arg68Ala, Lys69Ala and Arg106Ala decreases binding affinity to form the dimer 7·2-fold relative to wild-type nsp9, while Tyr82Ala enhance dimer stability, by a 8·0-fold increase in binding affinity (Z. Zeng et al. 2018). Additionally, Zeng et al. (2018), studied Porcine Epidemic Diarrhea Virus (PEDV) nsp9 residues involved in ssDNA binding activity and they identified five mutations that affected this process, namely, Lys10Ala, Arg68Ala, Lys69Ala, Arg106Ala and Tyr82Ala. The equivalents in SARS-CoV-2 identified from a sequence alignment are Arg10Ala, Arg74Ala, Arg111Ala, and Tyr87Ala, which are conserved in SARS-CoV-1.
Because SARS-CoV-1 and SARS-CoV-2 sequences are highly conserved (sequence identity is 97%), much structural information from SARS-CoV-1 is transferable to SARS-CoV-2. The three substitutions in SARS-CoV-2 nsp2 relative to SARS-CoV-1, Asn34Thr, Ser35Thr and His48Leu, are located on the nsp9 surface, distant from the dimerization region. Particularly, substitution Asn34Thr can result in an additional phosphorylation site, but phosphorylation of nsp9 has not yet been reported. As Thr34 and Thr35 are close to a potential ubiquitination site (Lys36), we hypothesize that their phosphorylation may prevent nsp9 ubiquitination.
