Nonstructural protein 4
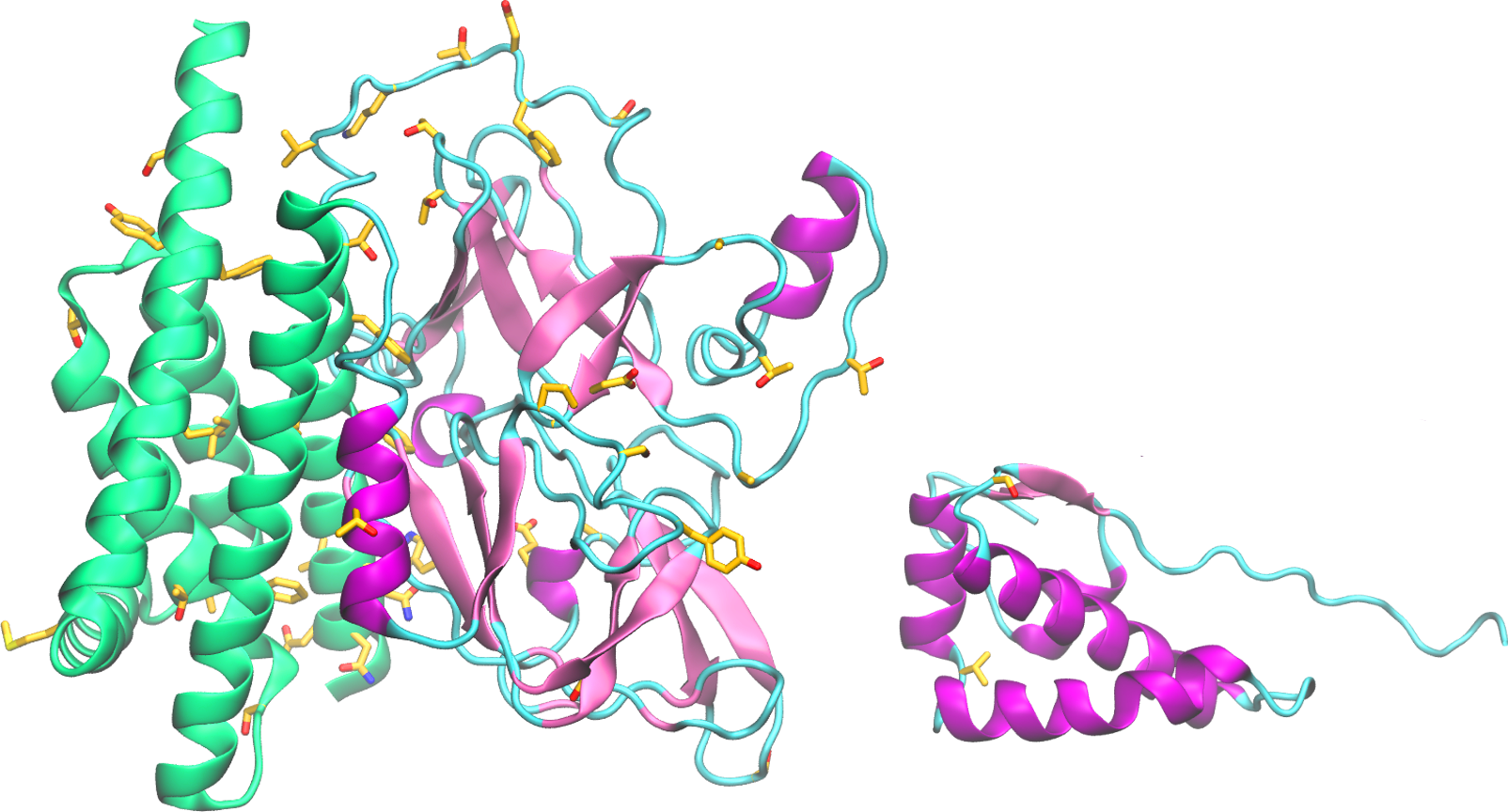
The nonstructural protein 4 (nsp4) is essential for membrane rearrangements during viral replication in a mechanism involving nsp3. Nsp4 is thought to be a tetra spanning transmembrane protein and disruption in glycosylation sites within the luminal loop give rise to aberrant double membrane vesicles. Structural information about nsp4 is scarce, thus a low-resolution model was generated for the full nsp4, using the ab initio protocol. The sequence of SARS-CoV-2 nsp4 is 80% identical to SARS-CoV-1 nsp4. However, non-conservative substitutions may affect the arrangement and packing of transmembrane segments.
Narrative
Working in coordination with nsp3 and nonstructural protein 6 (nsp6), the nonstructural protein 4 (nsp4) of SARS-CoV-1 is essential for membrane rearrangements during viral replication (Sakai et al. 2017; Angelini et al. 2013). Data suggests that coexpression of nsp3 with nsp4 results in host membrane rearrangement and the formation of double membrane vesicles and convoluted membranes (Hagemeijer et al. 2014). Nsp4 in mouse hepatitis virus, another coronavirus, is a glycosylated protein and is demonstrated to be involved in virally induced membrane rearrangement, replication complex assembly, and assembly of double membrane vesicles (Clementz et al. 2008; Gadlage et al. 2010; Oostra et al. 2007; Sparks, Lu, and Denison 2007; Beachboard et al. 2013). Aberrant double membrane vesicles and impaired RNA replication can be observed when mouse hepatitis virus nsp4 is lacking a glycosylation site (Beachboard, Anderson-Daniels, and Denison 2015; Gadlage et al. 2010). Prevention of interaction between nsp4 and nsp3 eliminated viral replication (Sakai et al. 2017).
Structural analysis and comparison with SARS-CoV-1 nsp4 – Nsp4 of SARS-CoV-1 is a nonstructural protein derived from the replicase polyprotein and is thought to be a tetra spanning transmembrane protein (Oostra et al. 2007). Disruption in glycosylation sites within the luminal loop between transmembrane domains 1 and 2 give rise to aberrant double membrane vesicles (Gadlage et al. 2010). Key amino acid residues His120 and Phe121 of nsp4 of SARS-CoV-1, which are conserved in SARS-CoV-2, are essential for interaction and binding of nsp4 to nsp3 as well as viral propagation (Sakai et al. 2017).
Structural information about nsp4 is scarce. A low-resolution model was generated for the full nsp4 using the ab initio protocol (figure, left). The soluble C-terminal domain is solved for a homologue (3vcb, MHV nsp4), enabling the generation of a higher resolution model for this region using the fragment-based modeling protocol (figure, right). Despite the likely inaccurate orientation of domains in the full protein model, the predicted secondary structures are consistent with the expected positions of the transmembrane and soluble domains. This model is a useful resource for general structural analysis.
The sequence of SARS-CoV-2 nsp4 is 80% identical to SARS-CoV-1 nsp4. Among the non-conservative substitutions, the predicted model reveals the substitution of four cysteines in the predicted TM helices by the bulky amino acids, Trp6, Phe7, Phe24 and Phe390. These substitutions may affect the arrangement and packing of transmembrane segments. Several substitutions are predicted to occur on the surface of the modeled soluble region spanning amino acids 33-275. The cytoplasmic C-terminal domain is highly conserved.
