Nonstructural protein 16 – 2’-O-ribose methyltransferase
The nonstructural protein 16 (nsp16) is involved in capping of viral mRNA to protect it from host degradation, and it has been demonstrated that it has to be associated with nsp10 to be active. Mutation that increases hydrophobic interaction between nsp10 and the hydrophobic pocket of nsp16, increases methyltransferase activity of nsp16. Conversely, a mutation in the RNA binding site of nsp16, completely abolished its methyltransferase activity. All these key amino acids are conserved in SARS-CoV-2 nsp16, but non-conservative substitutions in their vicinity may have a steric effect.
Narrative
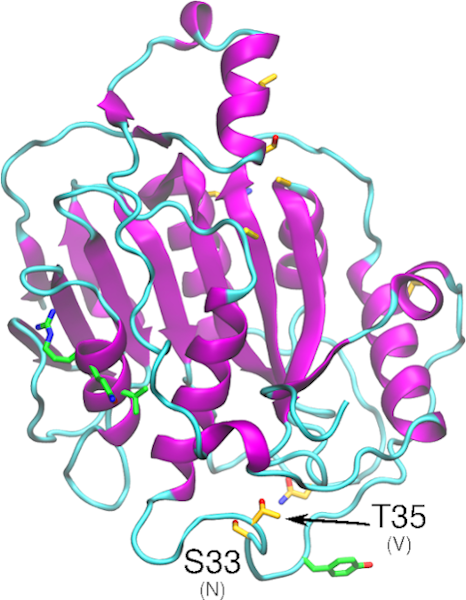
The nonstructural protein nsp16 is involved in capping of viral mRNA to protect it from host degradation, and it has been demonstrated that it has to be associated with nsp10 to be active. One potential method of ablating nsp16 activity is to disrupt the binding interface between nsp16 and nsp10. In SARS-CoV-1, the following interfacial mutations in nsp16 were shown to ablate nsp16 2’-O-methyltransferase activity: Ile40Ala, Met41Ala, Val44Ala, Val78Ala, Arg86Ala, Val104Gly, Leu244Ala and Met247Ala.(Decroly et al. 2011) In a separate study, a double mutant (His83Ala/Pro84Ala) and a triple mutant (Tyr76Ala/Cys77Ala/Arg78Ala) of nsp10 abolished SAM and m7GppA-RNA binding to nsp16 (Chen et al. 2011), further supporting the essential role of nsp10 in activating nsp16. In terms of the physicochemical interfacial properties that are essential for activating nsp16, it appears that certain hydrophobic interactions play a critical role and that enhancement of these may increase nsp16. This is exemplified by the Tyr96Phe mutation in nsp10 that increases methyltransferase activity by increasing hydrophobic interaction between nsp10 and the hydrophobic pocket of nsp16, which includes the sidechain of Val84 and the mainchain portions of Gln87 and Arg86 (Chen et al. 2011). In SARS-Cov-1, residue Tyr30 in the RNA binding site of nsp16, when mutated to either alanine or phenylalanine, abolished methlyltransferase activity, suggesting the critical role of this residue in RNA binding and, consequently, the enzymatic function (Decroly et al. 2011). All these key amino acids are conserved in SARS-CoV-2 nsp16, but non-conservative substitutions in the vicinity of Tyr30, namely Asn33Ser and Val35Thr may affect RNA binding, as well as conservative substitutions (Glu32Adp and Ile36Leu) that may have a steric effect.
