Nonstructural proteins 10 and 14 – replication and transcription
The exonuclease domain of nsp14 is imperative for replication fidelity within RNA viruses and has been shown to function as a proofreading exoribonuclease. Nsp14 associates with nsp10 that provides the ability to excise mismatched nucleotides, and disruption of this heterodimer was shown to decrease replication fidelity. SARS-CoV-1 and -CoV-21 nsp10 proteins are highly conserved (identity 97%), with no non-conservative substitutions between them. SARS-CoV-2 nsp14 is 73% identical to the SARS-CoV-1 counterpart. The substitution Glu128Pro in nsp14 is found close to the interface with nsp10, potentially affecting binding of nsp10-nsp14.
Narrative
Nsp10 is one of sixteen nonstructural proteins of the SARS-CoV-2 proteome. It interacts with nsp14 and nsp16 to perform 3’-5’ exoribonuclease and 2′-O-methyltransferase activities, respectively (Bouvet et al. 2010; Y. Wang et al. 2015). Disturbance in the interaction between nsp10 and nsp16 has been shown to be crippling to the virus, although still functional (Bouvet et al. 2014). Nsp14, nsp16, and nsp10 have been associated with the coronavirus wide replication and transcription complex (Sawicki et al. 2006, 2005). Crystal structures of SARS-CoV-1 nsp10-nsp16 have been reported (Decroly et al. 2011).
Structural analysis and comparison with SARS-CoV-1 nsp9 – SARS-CoV-1 and SARS-CoV-2 nsp10 proteins are highly conserved (identity 97%), with no non-conservative substitutions between them. These proteins are composed of 139 a.a. residues and fold forming two zinc fingers (Bouvet et al. 2014). This tight interaction with nsp14, involving multiple hydrogen bonds, salt bridges and hydrophobic packing, suggests that nsp10 might be necessary to maintain the integrity of nsp14 exoribonuclease domain (Ma et al. 2015; Bouvet et al. 2012).
The nonstructural protein nsp14 plays a critical role in coronaviruses replication and transcription. The exonuclease domain of nsp14 is imperative for replication fidelity within RNA viruses and has been shown to function as a proofreading exoribonuclease. Nsp14 associates with other nonstructural proteins from ORF1a and ORF1ab, including nsp7, nsp8, nsp12, and nsp10 (Ma et al. 2015; Subissi et al. 2014; Eckerle et al. 2007, 2010; Denison et al. 2011). The association with nsp10 provides the ability to excise mismatched nucleotides (Bouvet et al. 2012), and disruption of this heterodimer was shown to decrease replication fidelity (Smith et al. 2015).
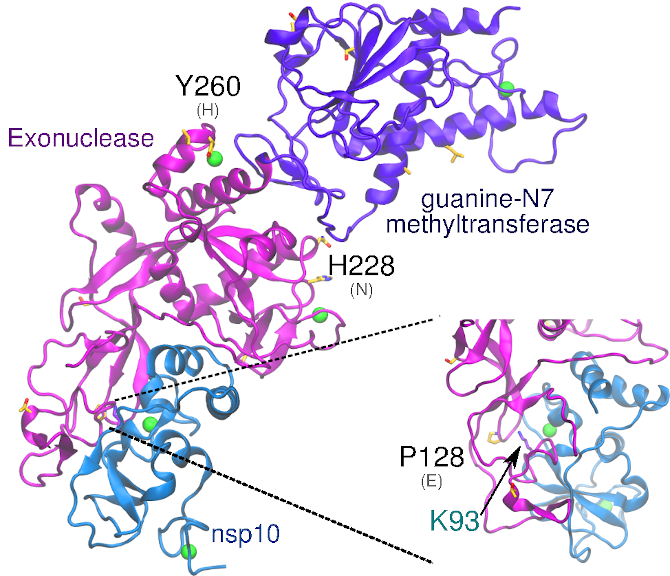
Structural analysis and comparison with SARS-CoV-1 nsp14 – Similar to SARS-CoV-1, SARS-CoV-2 nsp14 protein contains two catalytic domains: (1) The N-terminal exonuclease domain and (2) the C-terminal guanine-N7 methyltransferase domain. These domains, N-terminal and C-terminal, span a.a. sequences 1-287 and 288-527, respectively. The exonuclease domain also has conserved residues Asp, Glu, Asp and Asp (DEDD) that are key indicators of the DEDDh superfamily of DNA and RNA exonucleases.(Smith and Denison 2013; Zuo and Deutscher 2001) The guanine-N7 methyltransferase domain has been shown to be necessary for viral mRNA capping(Ma et al. 2015), hiding viral mRNA from host mRNA degradation machinery.
Nsp14 contains two zinc fingers within the N-terminal domain. The first is composed of Cys207, Cys210, Cys226 and His229, while the second is composed of His257, Cys261, His264, and Cys279 (Ma et al. 2015). Ala1-Arg76 and Ala119-Asp145 of nsp14 interact with nsp10. The third zinc finger of nsp14 is within the C-terminal domain and is formed by Cys452, Cys477, Cys484, and His487.(Ma et al. 2015)
SARS-CoV-2 nsp14 is 73% identical to the SARS-CoV-1 counterpart. Among the 14 non-conservative substitutions, Glu128Pro is the only substitution located close to the interface with nsp10. The proximity with the positively charged Lys93 of nsp10 in the heterodimer model suggests that this substitution may slightly affect binding of nsp10-nsp14. The substitutions His260Tyr and Asn228His may also be relevant as they are located in close proximity to the zinc fingers in the N-terminal exonuclease domain.
